Design of highly functional genome editors by modeling the universe of CRISPR-Cas sequences
Table Of Content
- Variation of existing structures
- Protein design articles within Nature Biotechnology
- SPDesign: protein sequence designer based on structural sequence profile using ultrafast shape recognition
- Efficient proximity labeling in living cells and organisms with TurboID
- Extended Data Fig. 8 Targeted unconditional and fold-conditioned protein binder design.
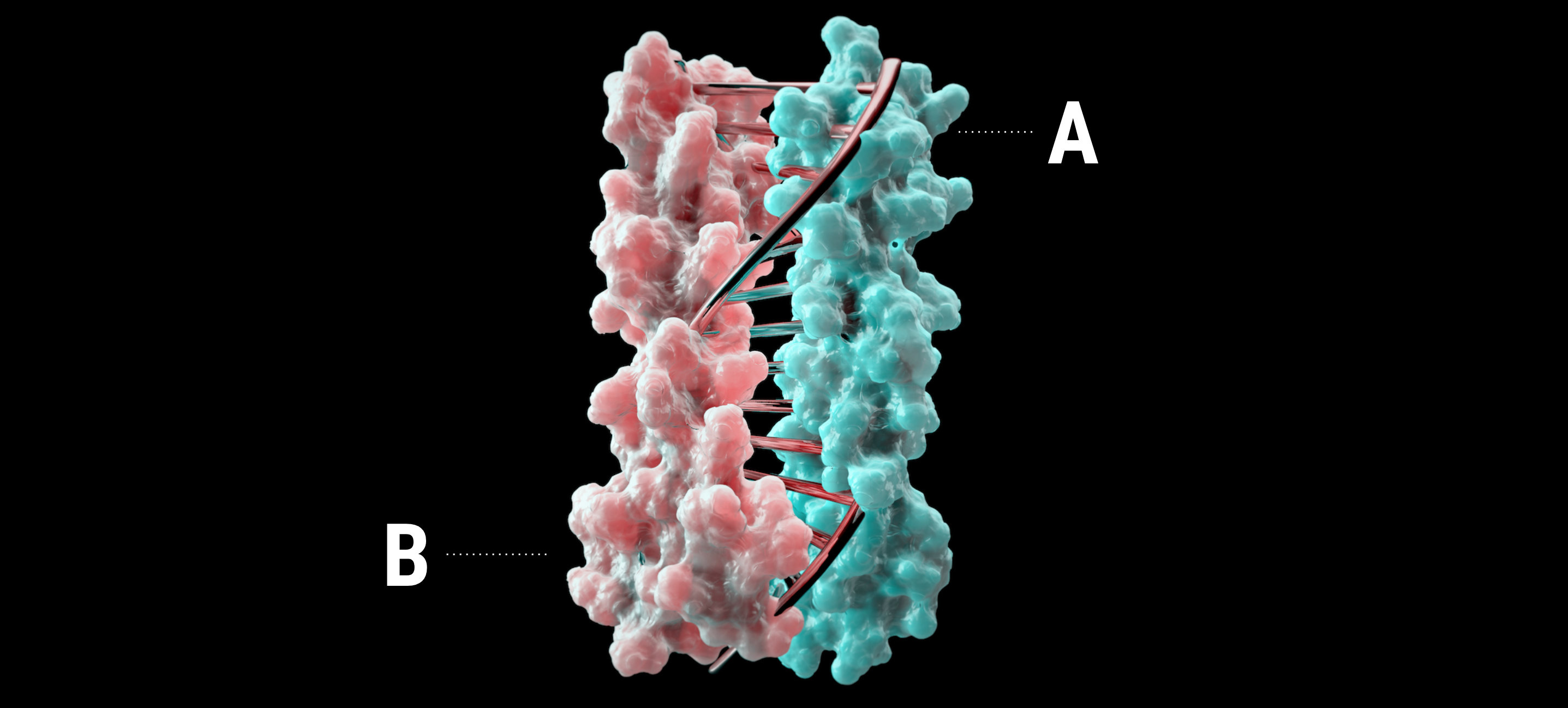
C, de novo proteins self-assemble into heterodimers (120), two-dimensional materials (9), filaments (8), cages (140), and alpha amyloids (143). D, a de novo–designed multipass transmembrane protein that has a defined membrane orientation (148). The designed DANCER protein has a tryptophan side chain that switches between predicted conformational states on the millisecond timescale (152). Engineered versions of fluorescent proteins, such as “split GFP” and “split dsRed” have also been developed to study protein–protein interactions in vivo66–68 (Figure 16). As the name implies, these assays use versions of fluorescent proteins that have been split into N- and C-terminal halves. Attaching proteins that bind to each other brings the two chains together, and the fluorescent protein is reconstituted.
Variation of existing structures
This contrasts with other forms of protein engineering, such as directed evolution, where a variety of methods are used to find proteins that achieve a specific function, and with protein structure prediction where the sequence is known, but the structure is unknown. Rational protein design approaches make protein-sequence predictions that will fold to specific structures. These predicted sequences can then be validated experimentally through methods such as peptide synthesis, site-directed mutagenesis, or artificial gene synthesis. Many protein functions involve interactions with other types of molecules such as DNA, RNA, saccharides, or small molecules.
Protein design articles within Nature Biotechnology
SEWING, structure extension with native-substructure graphs; TR, transform-restrained. Advances in side-chain design.A, in layer design, polar residues (cyan) are only allowed at surface and boundary positions, while hydrophobic residues (yellow) are only allowed at boundary and core positions. B, structures generated by side chain design methods can be evaluated by a set of filters, such as core packing quality, hydrogen bond satisfaction and local sequence/structure compatibility.
SPDesign: protein sequence designer based on structural sequence profile using ultrafast shape recognition
AI generates proteins with exceptional binding strength - ASBMB Today
AI generates proteins with exceptional binding strength.
Posted: Sat, 03 Feb 2024 08:00:00 GMT [source]
Raphael's model for evaluating protein structures is posted on bioRxiv as a preprint. The new method builds on the decades-long efforts of chemists to elucidate the three-dimensional structure of proteins and to use computers to search for suitable potential drug molecules. Until now, this has often involved laborious manual work, and in many cases the search yielded molecules that were very difficult or impossible to synthesise.
But existing gene and protein databases are constrained by a limited range of species and a heavy bias towards humans and commonly used model organisms. Basecamp Research is building an ultra-diverse repository of biological information obtained from samples collected in biomes in 17 countries, ranging from the Antarctic to the rainforest to hydrothermal vents on the ocean floor. Chief Technology Officer Philipp Lorenz says that once the genomic data from these specimens are analyzed and annotated, they can assemble a knowledge-graph that can reveal functional relationships between diverse proteins and pathways that would not be obvious purely on the basis of sequence-based analysis. “We are finding protein families in prokaryotes that have been thought to exist only in eukaryotes.” This means many more starting points for AI-guided protein design efforts, and Lorenz says that his team’s own design experiments have achieved an 80% success rate at producing functional proteins. This ML toolbox could generate made-to-order proteins too, including those with functions not present in nature. This is an appealing prospect because, despite natural proteins’ vast molecular diversity, there are many biomedical and industrial problems that evolution has never been compelled to solve.
By fusing domains from different proteins, it is possible to create novel networks that possess different input–output combinations. Interestingly, instead of forming the expected flat assembly based on the hexagonal design, the structures assembled into closed spheres (Figure 13, panels B and C). Modeling suggested that the hubs are actually wedge-shaped instead of perfect tripods with arms angled at exactly 120°. The largest reported assembly with structural validation, a 24-subunit protein cube, was designed by Yeates et al.58 Their design strategy involved making fusions between natural dimeric and trimeric proteins.
GenKIC was also applied to design meso-size proteins stabilized by multivalent cross-linkers (80). Despite these problems, there has been significant progress in developing computational techniques for predicting and designing protein structures de novo. This is in part because by examining “static” protein structures, designers can delineate design goals in a relatively straightforward manner. Certain elements can be designed for, such as “binding of the transition state” but the dynamics that accompany—and may be essential for—activity are not nearly so obvious. The connection between structure, dynamic protein–protein interactions and catalysis is not well understood. The goal of protein design is to find a protein sequence that will fold to a target structure.
By genetically encoding SpyTag and SpyCatcher in constructs of interest, diverse topologies can be created50 (Figure 9, panel B). Arnold, Tirrell and colleagues have exploited this technology for the construction of new protein-based materials.51 A covalent hydrogel network was produced as a result of isopeptide bond formation by genetically encoding SpyTag and Spy-Catcher into elastin-like protein (ELP) constructs (Figure 10). Moreover, chemistry allows researchers to explore the vast chemical space of possible amino acid sequences and develop strategies for the rational design of proteins with novel properties. Techniques such as directed evolution, which uses iterative rounds of mutation and selection to evolve proteins with enhanced functions, are rooted in the principles of chemical biology. On 26 January, Profluent came out of stealth mode with $9 million in seed funding to support the company’s efforts to apply machine learning (ML) to engineer novel functional proteins.
Extended Data Fig. 8 Targeted unconditional and fold-conditioned protein binder design.
The difference between the desired structure and the predicted structure can be backpropagated through the neural network to optimize the sequence (109) (blue). A, a membrane scoring function (124) uses a continuous hydration fraction to calculate the free energy change of residues from water to the lipid environment. B, protein design scoring functions are generalized to model small molecules (132) and carbohydrates (131). C, the TERMs-based scoring function (133) breaks proteins into tertiary structure motifs and evaluates the fitness of the sequence for any local structure using the sequence profiles of the tertiary motifs.
As a result, there is a great need in this industry for highly skilled workers and researchers. Nonetheless, the protein design and engineering market's expansion is hampered by the absence of such knowledge. Some of the greatest excitement in the deep learning world relates to generative models that can create entirely new proteins, never seen before in nature. These modeling tools belong to the same category of algorithms used to produce eerie and compelling AI-generated artworks in programs like Stable Diffusion or DALL-E 2 and text in programs like ChatGPT. In those cases, the software is trained on vast amounts of annotated image data and then uses those insights to produce new pictures in response to user queries.
Last January, Generate Biomedicines signed a $50 million drug development deal with Amgen that could potentially net the company more than $1.9 billion in total, and a few months later, Arzeda drew $33 million in series B funding to support its ongoing protein design programs. Other startups are also starting to crowd the field, such as computational company Cradle, which exited stealth in November with a $5.5 million seed investment, and Monod Bio, which launched with $25 million in seed funding in August. New small molecule-inducible domains that respond to novel stimuli would also be useful. To create more intricate synthetic pathways it will be necessary to develop additional switches that can be used to control the activity of multiple proteins independently while minimizing interference with native cellular processes.
The GenKIC method (76) adapted the robotics-inspired kinematic closure algorithm (77, 78) from loop modeling, generalized the approach to sample noncanonical backbone degrees of freedom, and applied it to cyclic peptides and peptides constrained by disulfide bonds. The designed peptides closely matched the experimentally solved structures and showed high stability against thermal and chemical denaturation. Kinematic closure methods in Rosetta (76, 78) can be used to enumerate backbones of cyclic peptides with seven to ten residues nearly exhaustively (79).
HBNet constructs a graph whose nodes are rotamers that have hydrogen bond donors or acceptors. Two nodes are connected by an edge if the rotamers of the nodes can form hydrogen bonds. HBNet was successfully applied to design helical bundle homo-oligomers with specificity mediated by hydrogen bond networks. A Monte Carlo version of the HBNet method uses a stochastic algorithm to traverse the HBNet graph (101). This new approach significantly improves the sampling speed and makes larger design problems possible. The ability to scaffold functional sites with any desired symmetry opens up new approaches to designing metal-coordinating protein assemblies49,50.
I want to show that this supplement really fills you up with energy unlike any other, just from the look while standing out from other similar products. I was given a brief of what information needed to go on both labels and I was also given the illustrations on the back directions. The client wanted a pouch design that will be simple also to include big logo and to have window so the consumer can se the liquid inside. Design is clean and modern and differentiates the product from the others in the market.
The success rates for studies where proteins were de novo–designed to have new structures are varied but can be high with many designs (blue). In contrast, success rates and numbers of successful designs for proteins with new functions (green) are much lower, except in a few cases where functional designs were all-helical proteins (red). Only studies that reported ten or more experimentally characterized designs (Table 1) are included.
Comments
Post a Comment